Plastic pollution is one of the most persistent environmental threats of our time. At SMCS-Psi Data Analytics Pvt. Ltd., we are advancing the frontier of plastic biodegradation using a powerful fusion of bioinformatics, structural modeling, and machine learning. Our latest innovation focuses on generating and validating novel plastic-degrading enzymes with the potential to revolutionize sustainable waste management. The Challenge: Biodegrading Plastic with Precision Most plastics, especially polyethylene terephthalate (PET), are resistant to natural degradation, accumulating in ecosystems for decades. Although a few bacterial species are known to degrade plastics, their enzymes often lack the efficiency or stability required for real-world applications. Our goal: Design a new enzyme with enhanced plastic-degrading activity, identify its microbial source, and confirm its structure-function integrity using computational structural biology tools. Sequence Discovery Using HMM We began by mining known PETase-like enzymes using Blast search and later using Hidden Markov Models (HMMs) to capture conserved sequence motifs associated with plastic degradation. Using this HMM profile, we scanned microbial sequence databases to generate and identify novel enzyme sequences that share evolutionary signatures with known plastic-degrading enzymes. Discovery Highlight Among the sequences generated, we discovered a novel bacterial species previously uncharacterized in this context. This organism encoded a candidate enzyme with significant HMM alignment to the PETase family, hinting at a functional plastic-degrading mechanism. Structural Validation: Ramachandran Plot & PROCHECK After generating the protein’s 3D structure using homology modeling, we performed comprehensive structural validation: Ramachandran Plot Analysis > Over 90% of residues fell within the most favored regions, confirming a well-folded and stereochemically acceptable protein structure. PROCHECK Residue Geometry Assessment > Using PROCHECK, we evaluated dihedral angles, bond lengths, and residue planarity. The model showed excellent stereochemical quality, comparable to high-resolution crystal structures. These validations ensured that our predicted enzyme was structurally sound and suitable for downstream functional evaluation. Molecular Dynamics (MD) Simulation To explore the structural stability and dynamics of the enzyme in a realistic environment, we ran Molecular Dynamics simulations over 100 ns. Key insights from MD analysis: The enzyme maintained a stable backbone RMSD, indicating overall structural stability. Active site residues remained conformationally intact, suggesting functional reliability during substrate binding. Temperature and pressure equilibration parameters remained within optimal thresholds, reinforcing the enzyme’s thermodynamic viability. From Sequence to Sustainability: Our Vision This study exemplifies how AI-driven sequence mining, when paired with advanced structural biology, can lead to tangible breakthroughs in bioremediation and green technology. The novel enzyme we’ve discovered is now a candidate for: In vitro PET plastic degradation testing Directed evolution to improve kinetics and substrate specificity Integration into bioengineered microbial consortia for waste treatment plants Join Our Mission for a Greener Future At SMCS-Psi, we are committed to transforming environmental challenges into opportunities using science, computation, and innovation. If you are a researcher, biotech startup, or environmental policymaker, we welcome collaborations to scale and commercialize such enzyme-based solutions.
Tackling Antibiotic Resistance: Toward a Universal Antibacterial Strategy
One of our most pressing research areas at SMCS-Psi focuses on addressing the growing challenge of antibiotic resistance, particularly in difficult-to-control plant and environmental pathogens such as Ralstonia solanacearum. This soil-borne bacterium is known for causing bacterial wilt in a wide range of crops, leading to significant agricultural losses worldwide. However, the broader implications of our research extend far beyond this single pathogen. Using our in-house machine learning platform, we have successfully screened over 10,000 natural and semi-synthetic compounds to identify potential antibacterial agents with high predicted efficacy. These molecules undergo a rigorous multi-step validation process that includes: Molecular Docking – Predicting the most favourable binding orientation of candidate compounds to essential bacterial proteins. LigPlot+ Interaction Visualization – Generating intuitive 2D diagrams that reveal key hydrogen bonds and hydrophobic interactions between the ligand and protein active site. Molecular Dynamics (MD) Simulations – Simulating real-time atomic-level interactions to evaluate the stability, flexibility, and binding free energy of the protein-ligand complex. In-vitro Biological Assays – Experimentally testing the top compounds to confirm their antimicrobial activity under laboratory conditions. Future Vision: A Universal Inhibitor Against Bacterial Growth Looking ahead, this computational-experimental hybrid pipeline is being extended to develop a broad-spectrum antibacterial compound—one capable of halting bacterial growth across multiple species by targeting a highly conserved and essential bacterial pathway. Our scientific rationale involves identifying evolutionarily conserved bacterial biomolecules, such as: DNA gyrase/topoisomerases Bacterial specific ribosomal proteins MurA (peptidoglycan biosynthesis) Fatty acid biosynthesis enzymes Two-component regulatory systems These proteins are indispensable for bacterial survival and present minimal variation across Gram-positive and Gram-negative strains. By using ML-based comparative proteomics and ligand-based activity prediction, we aim to pinpoint lead molecules that bind to these targets with: High selectivity Minimal off-target toxicity Resistance-resilient binding mechanisms Machine Learning as the Driver Our ML framework is continuously being trained on curated datasets of: Known antibiotic–protein interaction profiles Resistance gene patterns (from databases like CARD and ResFinder) Multi-target inhibitors Physicochemical & pharmacokinetic properties This enables prediction of both antibacterial activity and resistance risk, allowing us to prioritise compounds that are both potent and durable. Additionally, explainable AI techniques (e.g., SHAP, integrated gradients) help us identify which molecular substructures contribute most to predicted activity—guiding rational design and chemical optimisation. Broader Impact This initiative doesn’t just aim to combat Ralstonia infections in plants but also contributes toward: Developing agriculturally safe antimicrobials Designing next-generation antibiotics for clinical pathogens Building a generalisable ML framework for drug discovery in infectious diseases In the long run, our goal is to create a first-in-class compound that can suppress or eliminate bacterial growth across diverse taxa by acting on a universal, irreplaceable bacterial function—a major milestone in global health and food security.
Accelerating Drug Discovery with Machine and Deep Learning at SMCS-Psi
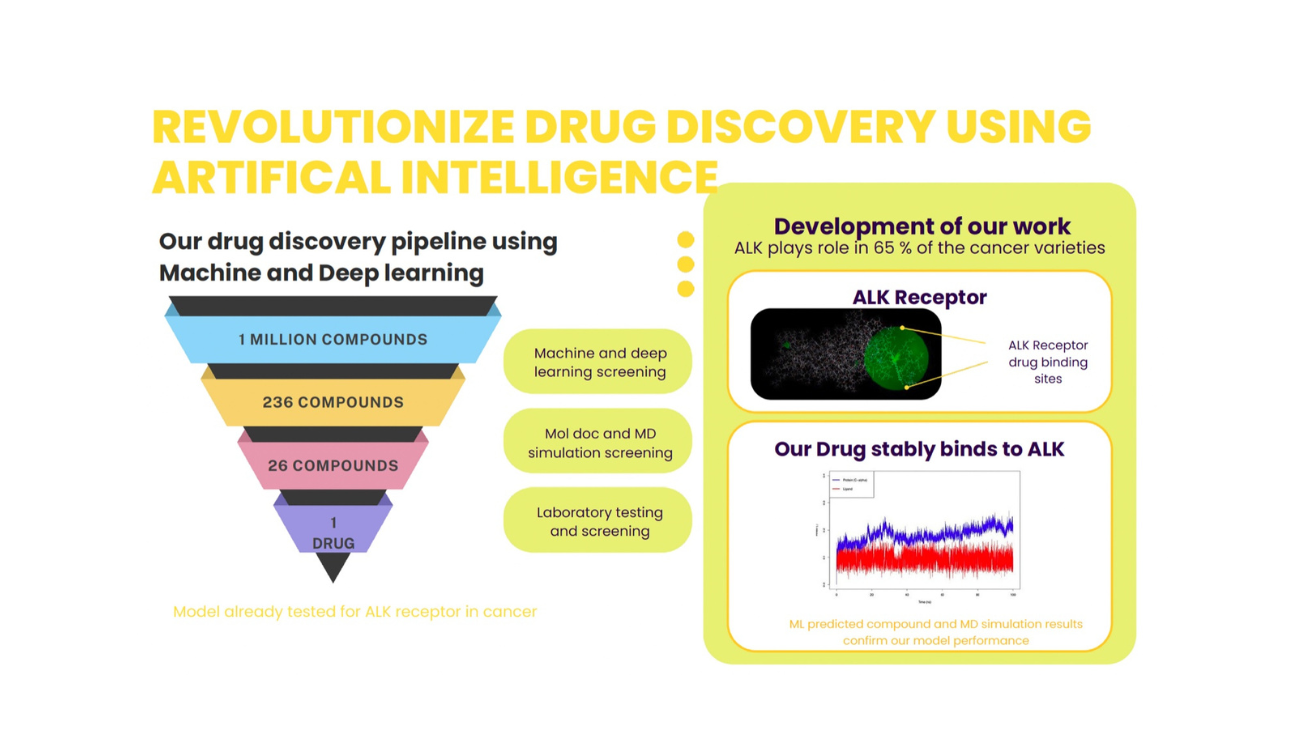
Posted by SMCS-Psi Data Analytics Pvt. Ltd. | Pune & Leipzig In the rapidly evolving world of pharmaceuticals, traditional drug discovery methods are often time-consuming, costly, and limited in scope. At SMCS-Psi Data Analytics Pvt. Ltd., we are redefining this process through the power of machine learning (ML) and artificial intelligence (AI). Our latest initiative brings together computational biology, cheminformatics, and deep learning to transform how new therapeutic compounds are identified, evaluated, and optimized. The Vision: Smarter, Faster Drug Discovery Drug discovery is a complex and multi-stage journey—from identifying biological targets and screening compounds to assessing binding affinities, and finally to testing efficacy and safety. Our team at SMCS-Psi is harnessing ML to automate and enhance these critical stages, reducing the development timeline and increasing the success rate of potential drug candidates. Machine Learning in Action: Our Approach At the core of our project lies a sophisticated ML-powered drug discovery pipeline designed to predict biological activity, optimize molecular properties, and simulate protein-ligand interactions. Key Components of Our ML Drug Discovery Pipeline Molecular Representation We convert chemical structures into SMILES strings and molecular graphs, enabling our models to digitally process and analyze thousands of compounds efficiently. Deep Learning for Activity Prediction Using advanced models like Graph Neural Networks (GNNs) and Recurrent Neural Networks (RNNs), we predict binding affinities, toxicity profiles, and ADMET properties of candidate molecules. Virtual Screening & Docking ML accelerates high-throughput screening, narrowing down top candidates for structure-based docking against validated biological targets. Target Identification By integrating genomic, proteomic, and gene expression data, we identify novel and high-impact therapeutic targets, especially those involved in complex diseases and resistance pathways. LigPlot Interaction Diagrams We use LigPlot+ to visualize detailed protein-ligand interactions post-docking. This offers a 2D schematic view of hydrogen bonds and hydrophobic contacts, allowing researchers to interpret binding modes clearly and refine lead compounds. Molecular Dynamics (MD) Simulations To ensure stability and validate the biological relevance of docked complexes, we perform MD simulations. These simulations help assess the conformational flexibility, binding energy, and interaction dynamics of protein-ligand complexes over time in realistic environments. Join Us in Revolutionising Drug Discovery At SMCS-Psi, we believe the future of medicine is data-driven, fast-tracked, and intelligent. If you’re a researcher, biotech entrepreneur, or pharma innovator looking to accelerate your R&D, we welcome collaborations and partnerships. Contact us today to learn more about our machine learning and simulation solutions for drug discovery—or to explore how we can support your next breakthrough. SMCS-Psi Data Analytics Pvt. Ltd. Where Biology Meets Data Science. Ravet, Pune (India) | Leipzig (Germany) info@smcs-psi.com www.smcs-psi.com